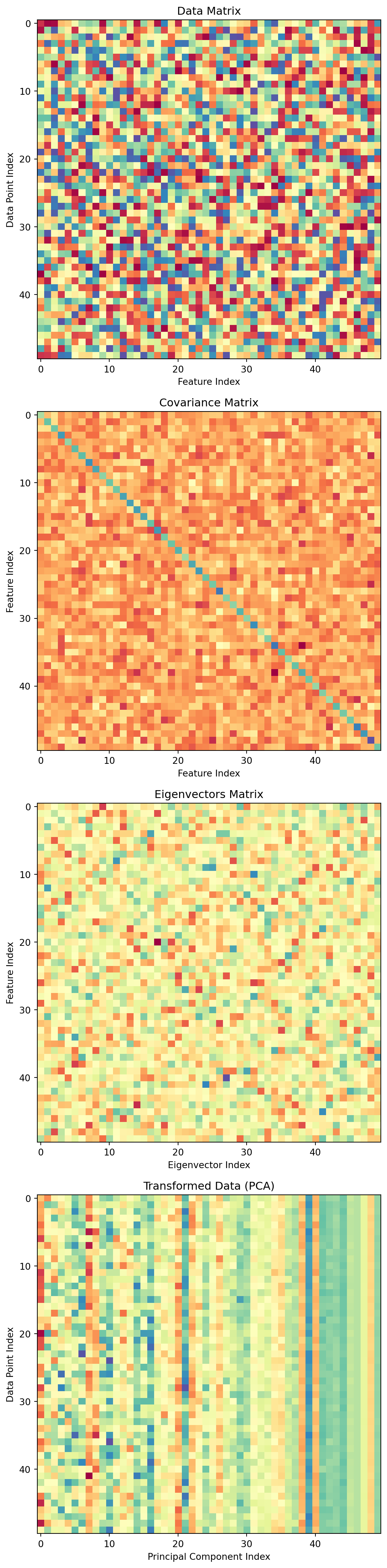
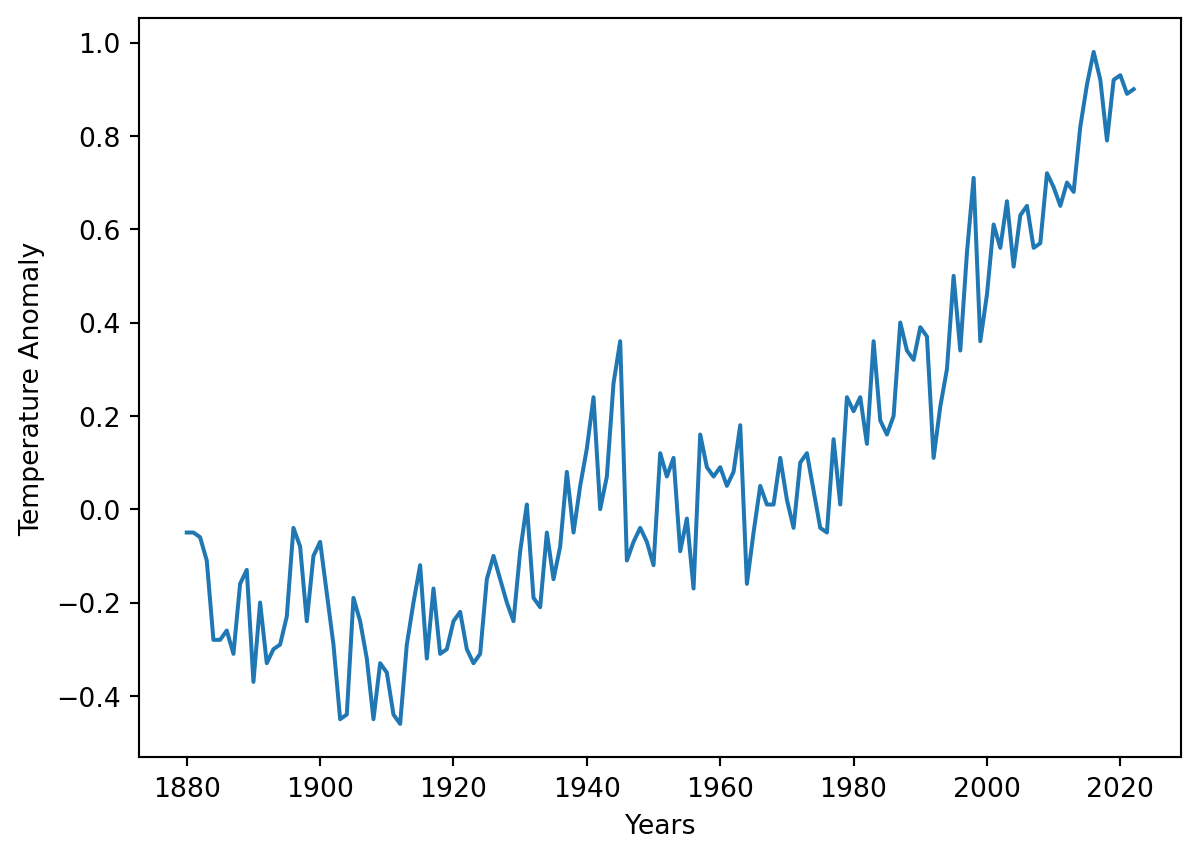
array([[ 1.880e+03, -5.000e-02],
[ 1.881e+03, -5.000e-02],
[ 1.882e+03, -6.000e-02],
[ 1.883e+03, -1.100e-01],
[ 1.884e+03, -2.800e-01],
[ 1.885e+03, -2.800e-01],
[ 1.886e+03, -2.600e-01],
[ 1.887e+03, -3.100e-01],
[ 1.888e+03, -1.600e-01],
[ 1.889e+03, -1.300e-01],
[ 1.890e+03, -3.700e-01],
[ 1.891e+03, -2.000e-01],
[ 1.892e+03, -3.300e-01],
[ 1.893e+03, -3.000e-01],
[ 1.894e+03, -2.900e-01],
[ 1.895e+03, -2.300e-01],
[ 1.896e+03, -4.000e-02],
[ 1.897e+03, -8.000e-02],
[ 1.898e+03, -2.400e-01],
[ 1.899e+03, -1.000e-01],
[ 1.900e+03, -7.000e-02],
[ 1.901e+03, -1.800e-01],
[ 1.902e+03, -2.900e-01],
[ 1.903e+03, -4.500e-01],
[ 1.904e+03, -4.400e-01],
[ 1.905e+03, -1.900e-01],
[ 1.906e+03, -2.400e-01],
[ 1.907e+03, -3.200e-01],
[ 1.908e+03, -4.500e-01],
[ 1.909e+03, -3.300e-01],
[ 1.910e+03, -3.500e-01],
[ 1.911e+03, -4.400e-01],
[ 1.912e+03, -4.600e-01],
[ 1.913e+03, -2.900e-01],
[ 1.914e+03, -2.000e-01],
[ 1.915e+03, -1.200e-01],
[ 1.916e+03, -3.200e-01],
[ 1.917e+03, -1.700e-01],
[ 1.918e+03, -3.100e-01],
[ 1.919e+03, -3.000e-01],
[ 1.920e+03, -2.400e-01],
[ 1.921e+03, -2.200e-01],
[ 1.922e+03, -3.000e-01],
[ 1.923e+03, -3.300e-01],
[ 1.924e+03, -3.100e-01],
[ 1.925e+03, -1.500e-01],
[ 1.926e+03, -1.000e-01],
[ 1.927e+03, -1.500e-01],
[ 1.928e+03, -2.000e-01],
[ 1.929e+03, -2.400e-01],
[ 1.930e+03, -9.000e-02],
[ 1.931e+03, 1.000e-02],
[ 1.932e+03, -1.900e-01],
[ 1.933e+03, -2.100e-01],
[ 1.934e+03, -5.000e-02],
[ 1.935e+03, -1.500e-01],
[ 1.936e+03, -8.000e-02],
[ 1.937e+03, 8.000e-02],
[ 1.938e+03, -5.000e-02],
[ 1.939e+03, 5.000e-02],
[ 1.940e+03, 1.300e-01],
[ 1.941e+03, 2.400e-01],
[ 1.942e+03, 0.000e+00],
[ 1.943e+03, 7.000e-02],
[ 1.944e+03, 2.700e-01],
[ 1.945e+03, 3.600e-01],
[ 1.946e+03, -1.100e-01],
[ 1.947e+03, -7.000e-02],
[ 1.948e+03, -4.000e-02],
[ 1.949e+03, -7.000e-02],
[ 1.950e+03, -1.200e-01],
[ 1.951e+03, 1.200e-01],
[ 1.952e+03, 7.000e-02],
[ 1.953e+03, 1.100e-01],
[ 1.954e+03, -9.000e-02],
[ 1.955e+03, -2.000e-02],
[ 1.956e+03, -1.700e-01],
[ 1.957e+03, 1.600e-01],
[ 1.958e+03, 9.000e-02],
[ 1.959e+03, 7.000e-02],
[ 1.960e+03, 9.000e-02],
[ 1.961e+03, 5.000e-02],
[ 1.962e+03, 8.000e-02],
[ 1.963e+03, 1.800e-01],
[ 1.964e+03, -1.600e-01],
[ 1.965e+03, -5.000e-02],
[ 1.966e+03, 5.000e-02],
[ 1.967e+03, 1.000e-02],
[ 1.968e+03, 1.000e-02],
[ 1.969e+03, 1.100e-01],
[ 1.970e+03, 2.000e-02],
[ 1.971e+03, -4.000e-02],
[ 1.972e+03, 1.000e-01],
[ 1.973e+03, 1.200e-01],
[ 1.974e+03, 4.000e-02],
[ 1.975e+03, -4.000e-02],
[ 1.976e+03, -5.000e-02],
[ 1.977e+03, 1.500e-01],
[ 1.978e+03, 1.000e-02],
[ 1.979e+03, 2.400e-01],
[ 1.980e+03, 2.100e-01],
[ 1.981e+03, 2.400e-01],
[ 1.982e+03, 1.400e-01],
[ 1.983e+03, 3.600e-01],
[ 1.984e+03, 1.900e-01],
[ 1.985e+03, 1.600e-01],
[ 1.986e+03, 2.000e-01],
[ 1.987e+03, 4.000e-01],
[ 1.988e+03, 3.400e-01],
[ 1.989e+03, 3.200e-01],
[ 1.990e+03, 3.900e-01],
[ 1.991e+03, 3.700e-01],
[ 1.992e+03, 1.100e-01],
[ 1.993e+03, 2.200e-01],
[ 1.994e+03, 3.000e-01],
[ 1.995e+03, 5.000e-01],
[ 1.996e+03, 3.400e-01],
[ 1.997e+03, 5.500e-01],
[ 1.998e+03, 7.100e-01],
[ 1.999e+03, 3.600e-01],
[ 2.000e+03, 4.600e-01],
[ 2.001e+03, 6.100e-01],
[ 2.002e+03, 5.600e-01],
[ 2.003e+03, 6.600e-01],
[ 2.004e+03, 5.200e-01],
[ 2.005e+03, 6.300e-01],
[ 2.006e+03, 6.500e-01],
[ 2.007e+03, 5.600e-01],
[ 2.008e+03, 5.700e-01],
[ 2.009e+03, 7.200e-01],
[ 2.010e+03, 6.900e-01],
[ 2.011e+03, 6.500e-01],
[ 2.012e+03, 7.000e-01],
[ 2.013e+03, 6.800e-01],
[ 2.014e+03, 8.200e-01],
[ 2.015e+03, 9.100e-01],
[ 2.016e+03, 9.800e-01],
[ 2.017e+03, 9.200e-01],
[ 2.018e+03, 7.900e-01],
[ 2.019e+03, 9.200e-01],
[ 2.020e+03, 9.300e-01],
[ 2.021e+03, 8.900e-01],
[ 2.022e+03, 9.000e-01]])